World Antimicrobial Awareness Week 2022: Preventing Antimicrobial Resistance Together
Spread Awareness, Stop Resistance
- World Antimicrobial Awareness Week
- Antimicrobial resistance (AMR)
- Priority pathogens
- Integrated stewardship: Collaboration with clinical laboratories to optimise antimicrobial therapy
- Genetic Signatures 3-base™ EasyScreen™ Detection Kit for AMR testing
- Evaluation Studies
World Antimicrobial Awareness Week
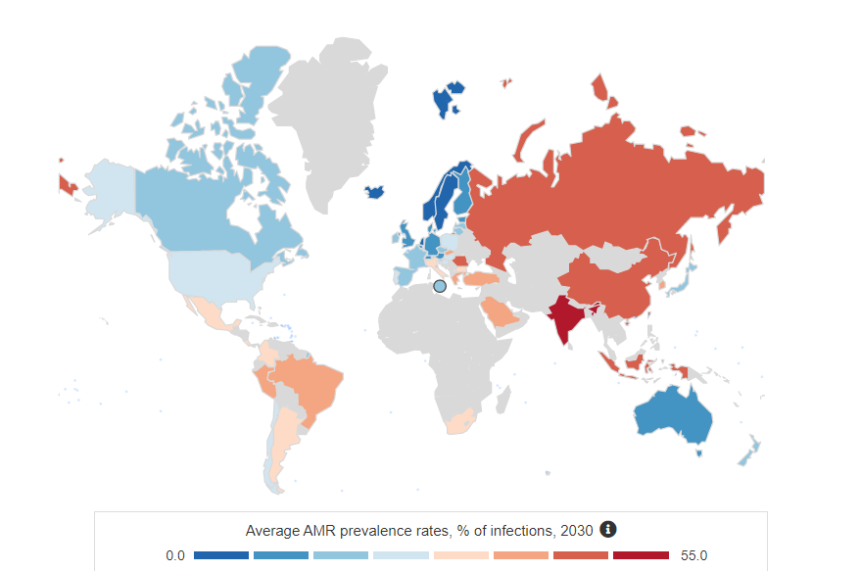
The World Health Organisation (WHO) has declared antimicrobial resistance (AMR) as one the most challenging threats to health. Global AMR prevalence rates are expected to continue to climb (Figure 1), with an estimated death toll projected to lift to 10 million by 2050 [1], pushing AMR-related deaths higher than cancer.
World Antimicrobial Awareness Week (WAAW) runs from the 18-24th November every year and seeks to improve knowledge about AMR and reduce the impact of this “silent pandemic”. WAAW highlights the global efforts to advance research, diagnostics, and patient care.
This year, the WAAW theme is Preventing Antimicrobial Resistance Together, calling stakeholders, policymakers, health care providers, and the general public to Go Blue for AMR. Genetic Signatures is on board, with unique and rapid diagnostic solutions to improve patient outcomes and save lives.
Figure 1: OECD projected global average AMR prevalence rates by 2030 [2].
Whilst these projections are concerning, AMR prevalence is expected to be even higher, with infection rates underrepresented to varying degrees across the globe, due to limited laboratory-based AMR surveillance and reporting. To identify, track and manage AMR burden, improved AMR surveillance through diagnostic testing is essential across health care, public, and environmental settings.
Antimicrobial resistance (AMR)
AMR is mainly caused by the overuse or misuse of antimicrobials (such as antibiotics), which has led to an increase in drug resistant pathogens worldwide. In response to antimicrobial exposure, microbes have evolved mechanisms to protect themselves, usually through genetic changes by spontaneous gene mutations, or acquisition of resistance genes from other bacteria [3]. These genetic changes can also increase the likelihood of resistance spread which can occur between people in the community, health care settings, in agriculture, and the environment. Resistance to antibiotic treatment has been recorded as early as the 1940s, in line with the first mass production of penicillin [4].
Bacteria resistant to specific antibiotics now cause almost one in five infections in OECD and EU28 countries [5]. Treatment of AMR microbial infections is becoming much more difficult, resulting in increased severity of the infection, prolonged illness, longer hospital stays, and potentially disability, and death. Hence, the health, social and economic burden of AMR is significant.
Microbes resistant to multiple antimicrobials are called multidrug resistant (MDR). There is increasing evidence of a growing spread of MDR bacteria that cause common infections such as urinary tract, sexually transmitted and gastrointestinal infections, and sepsis [6]. The spread of pan-resistant bacteria that are not treatable with any existing antimicrobial medicines (known as ‘super bugs’) is particularly concerning [7].
New antimicrobials are urgently needed – though the number entering the market has decreased significantly over the past decade, and there are few candidates in the drug development pipeline [2, 8]. In addition, if the community and health industry does not adjust how antibiotics are used, newly developed antibiotics will also become ineffective. Therefore, drug development alone is not the solution.
Priority pathogens
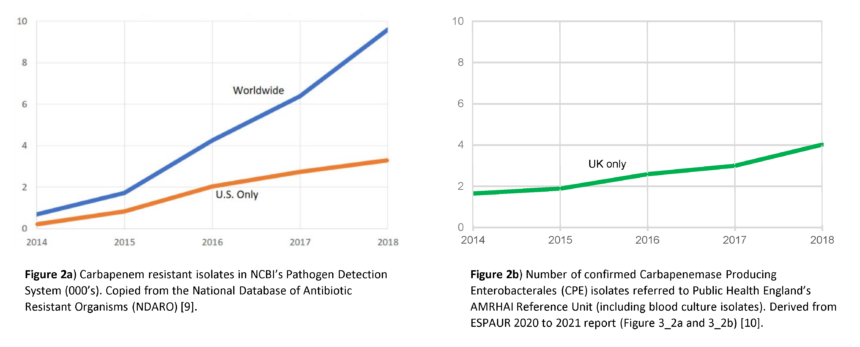
Beta-lactam and carbapenem antibiotics are most commonly used in the treatment of bacterial infections. However, the recent emergence of Extended-Spectrum Beta-Lactamases (ESBL) and Carbapenemase Producing Organisms (CPO) are a significant global concern, as these enzymes may render antibiotic treatment ineffective (Figure 2) [9, 10].
Figure 2. The estimated rise in CPO resistance isolates and markers worldwide, US and UK [9, 10].
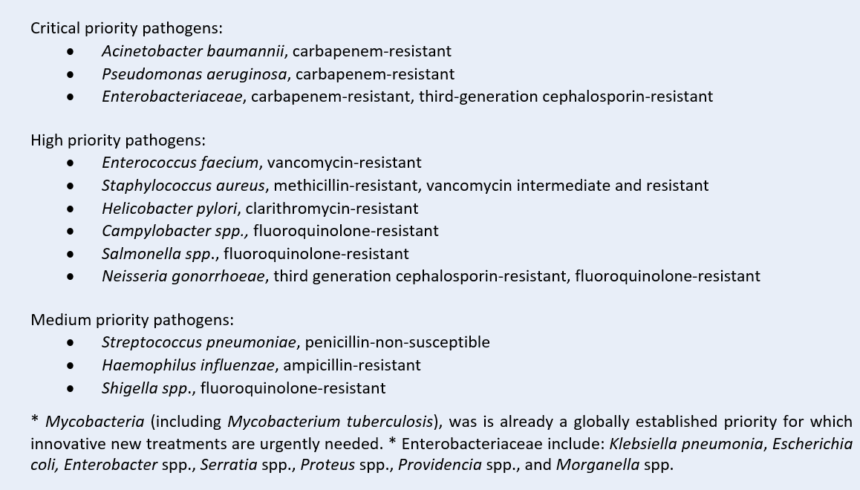
In 2017 the WHO published a global priority list of antibiotic resistant pathogens requiring urgent action (Figure 3). This list was divided into critical, high, and medium priority groups, according to the threat level each poses to human health [11]. Since this list was published, resistance has continued to grow, and will grow further unless action is taken. The CDC also publish an AR Threat Report [12], citing similar pathogens of concern, though adds Candida auris and Clostridioides difficile as an urgent threat. These additions are also similar for Australia (CARAlert) [13, 14] and the United Kingdom [15].
Figure 3: WHO global Priority Pathogens [11].
Integrated stewardship: Collaboration with clinical laboratories to optimise antimicrobial therapy
The importance of the correct management and use of antimicrobial drugs is essential to help preserve the effectiveness of these vital medical treatments. AMR infections are particularly common and problematic in hospitals and immunocompromised patients, and those undergoing surgery, are at higher risk. Medical procedures will pose further risks as AMR infections continue to grow, with repercussions of significant illness and mortality.
Integration of diagnostic testing through diagnostic stewardship, in partnership with antimicrobial stewardship programs, has led to significant improvements in the treatment and management of patients with infectious diseases. A multidisciplinary approach to these stewardship programs has decreased time to effective and optimal therapy, along with reducing mortality, length of hospital stay, and healthcare costs across various settings and patient populations [16-19] (Figure 4).
Figure 4: The relationship between diagnostic stewardship and antimicrobial stewardship [20, 21].
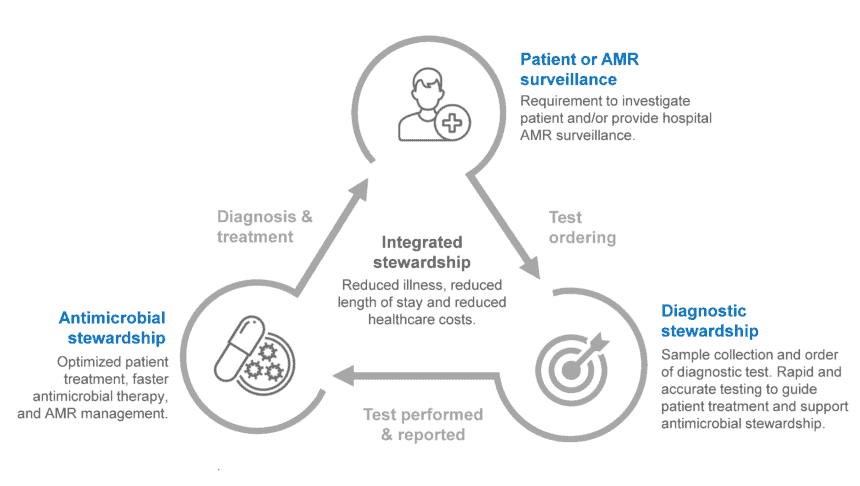
Diagnostic stewardship is defined by the WHO as a “coordinated guidance and interventions to improve appropriate use of microbiological diagnostics to guide therapeutic decisions. It should promote appropriate, timely diagnostic testing, including specimen collection, and pathogen identification and accurate, timely reporting of results to guide patient treatment.” [22]
Diagnostic stewardship can occur at various stages in a diagnostic test life cycle, with even minor adjustments to patient care offering significant benefits to both health and economic outcomes. A recent study [23] described key components essential for excellent delivery of patient care, listed below.
-
- Diagnostic intelligence: Diagnostic stewardship, decision support and risk stratification.
- Diagnostic readiness: Diagnostic subject matter expertise and value-based care.
- Diagnostic intervention: Diagnostic speed and accessibility.
- Diagnostic impact: Antimicrobial stewardship, radiologic stewardship, bed availability, general resource recovery, safety, quality and financial stewardship.
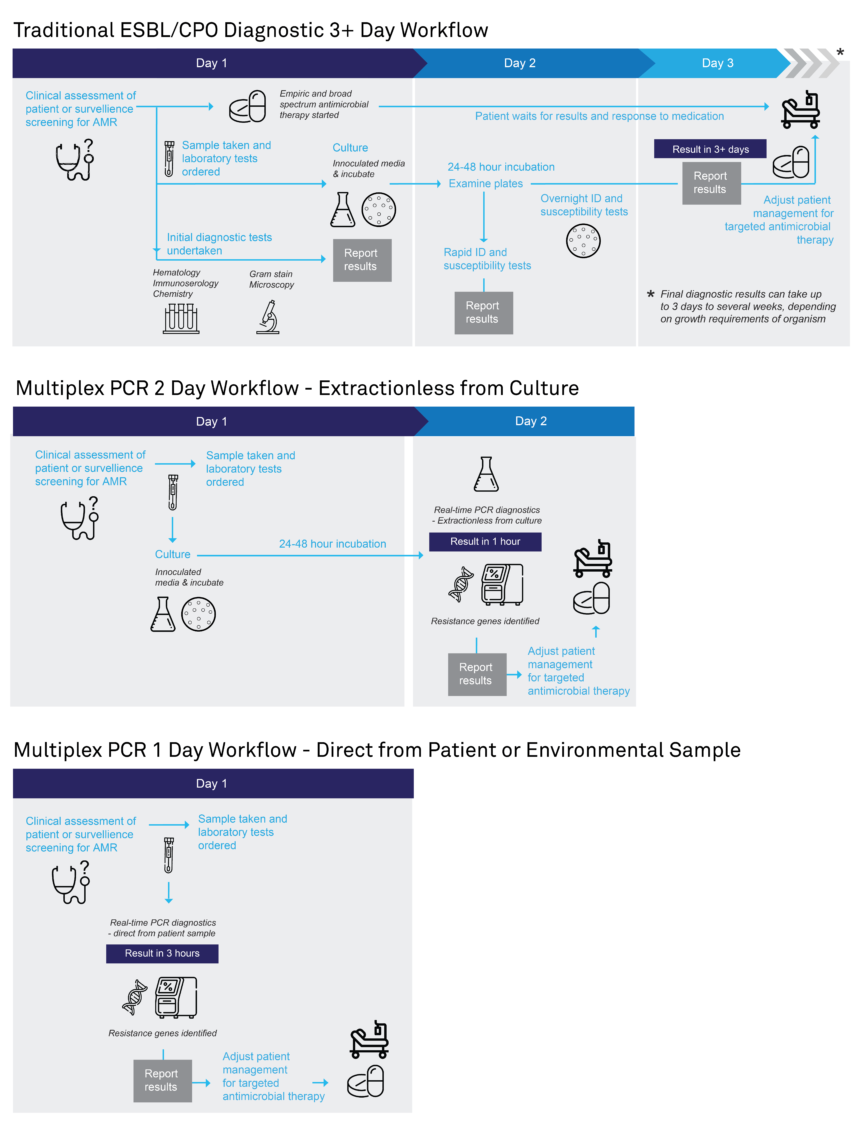
Rapid diagnostic testing offers significant advantages over conventional AMR diagnostics. Conventional AMR diagnostics uses antibiotic disk diffusion methods to assess antibiotic susceptibility on culture media. As multiple microbial culture processes are required, the time from sample to result is around 40+ hours. This process is tedious and distinguishing infections requiring antibiotics from self-limiting infections can be clinically challenging. In addition, longer turnaround times can delay the correct therapeutic treatment (Figure 5).
Of significant global concern is the emergence of ESBL and CPO associated infection in healthcare settings, as standard treatments may be rendered ineffective. The urgency of accurate and timely detection of these organisms is even more important, as delayed diagnosis and treatment could have a significant impact on patient health.
To address these challenges, demand for rapid diagnostic technology offering accurate detection of ESBL and CPO infections is growing. Comprehensive surveillance of AMR bacteria in hospital settings has been suggested as key methods to uncover the true prevalence and incidence of AMR, and in turn, provide the necessary information to control transmission, reduce outbreaks and the economic burden of ESBL and CPO infections [24].
Genetic Signatures are specialists in molecular diagnostics, providing a unique, rapid multiplex PCR-based assay for detection of the most significant and commonly encountered ESBL and CPO resistance genes.
Genetic Signatures 3base™ EasyScreen™ Assays for rapid and specific AMR diagnosis
Rapid, high throughput diagnostic screening solutions supporting antimicrobial stewardship procedures
Genetic Signatures is a specialist molecular diagnostics company, providing advanced syndromic molecular screening solutions for infectious disease for over 20 years. Genetic Signatures innovative diagnostic solutions is supported by using proprietary 3baseTM technology, simplifying molecular pathology and improving patient care.
Genetic Signatures’ EasyScreen™ ESBL/CPO Detection Kit has been developed as a useful tool to rapidly screen resistant genes associated with ESBL and COP infections. The novel 3base™ real-time multiplex-PCR assay can screen the most significant and commonly encountered bacterial resistance genes including: TEM, CTX-M, SME, GES, IMP, NDM, OXA-23 like, OXA-48 like, OXA-51 like, MCR-1, DHA, SHV, VIM, IMI, CMY, KPC and their subtypes.
The 3base™ assay works by specifically modifying the 4-base wild-type DNA sequence (A, C, T, G) into a 3-base sequence (A, T, G) via a novel, patented 3base™ conversion step. The conversion process simplifies the design of multiplex PCR reactions by eliminating the large Tm differences that can be present when targeting multiple pathogens, and increases the homology between sequences (Table 2). The increased sub-type homology of the 3base™ technique expands the detection capacity of multiplex-PCR for some target genes (example, Pan-CTX-M). Also, novel variants (such as IMP-14) or new resistant markers can be readily incorporated into existing assays given the properties of the 3baseTM converted DNA sequences, thus improving the throughput of such assays.
Table 1. Example of the 3base™ mechanism. The example sequence above demonstrates the increase in homology from 75% (before) to 95% (after) via the 3base™ conversion where all C bases are detected as T bases. Click here for more information on 3base™ technology.
The EasyScreen™ ESBL/CPO Detection Kit provides a sensitive and rapid alternative for the detection of ESBL and CPO sequences which can be carried out in less than 3 hours for primary patient specimens, and in approximately 1 hour for cultured isolates (Figure 5).
For cultured isolates, samples can be taken directly from cultures and broths without the need for DNA extraction/purification steps, significantly shortening turnaround times to result. The short turn around-time of 1 hour could be effective in assisting and accelerating patient management strategies.
Figure 5. Diagnostic pathway of convention AMR diagnostic methods compared to using Genetic Signatures EasyScreen™ ESBL/CPO Detection Kit. Click to download expandable AMR workflow.
Rapid screening of AMR resistance genes allows:
- The appropriate use of antimicrobial therapy
- Improved health outcomes for the patient
- Reduced length of hospital stay
- Significant savings for the patient and hospital
- Improved AMR detection and management
- Subsequent reduction in AMR outbreaks
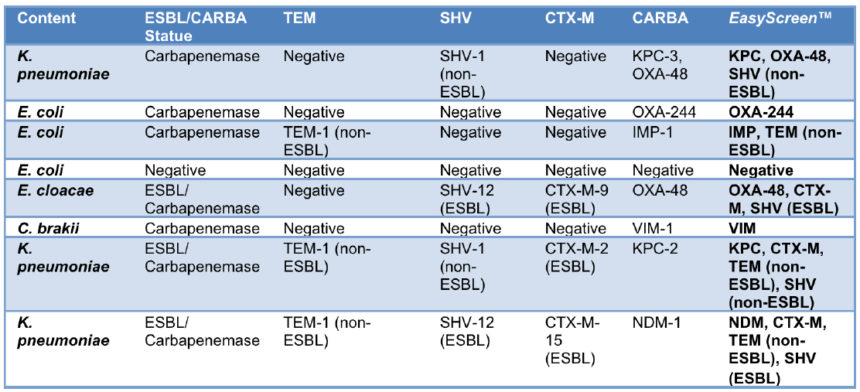
A recent clinical validation study undertaken at the National CPE Reference Laboratory in Galway, Ireland (ref to poster) (Table 3), found that two-thirds of the clinical isolates tested had a mix of infections. Multiplex PCR detection using the EasyScreen™ ESBL/CPO Detection Kit assisted in identifying additional EMBL/CPO markers missed by using conventional culture methods.
Evaluation Studies
Evaluation of the EasyScreen™ ESBL/CPO Detection Kit for the Detection of ß-Lactam Resistance Genes
The EasyScreen™ ESBL/CPO Detection Kit was recently evaluated for the molecular detection of ESBL and CPO targetson 341 bacterial isolates, assessing the benefit of employing molecular-based testing to traditional culture techniques [25]. The assay was found to be an efficient and rapid test (<3hrs) for the most frequently encountered penicillinases (including ESBLs), cephalosporinases, and carbapenemases, including IMP variants that are not well detected by most other molecular assays [26]. The results also demonstrated the assay is well adapted to the European epidemiology of Carbapenemase-Producing Enterobacterales and P. aeruginosa. Furthermore, the author noted the assay is adapted for the detection of the most prevalent carbapenemases encountered in Gram-negative bacteria isolated worldwide.
Genetic Signatures thanks the team who lead this study – Camille Gonzalez, Saoussen Oueslati , Laura Biez, Laurent Dortet and Thierry Naas. This study was in collaboration with the Université Paris-Saclay, Greater Paris University Hospitals – AP-HP and French National Reference Center for Antibiotic Resistance.
Evaluation of EasyScreenTM ESBL/CPO Detection Kit Using Direct-PCR from Patient Culture and Broth Samples
An additional evaluation study compared the performance of the EasyScreen™ ESBL/CPO Detection Kit using two workflow methods to detect ESBL and CPO resistance genes. The first approach employed Genetic Signatures’ automated extraction instrument, the GS-mini, on known reference material and clinical isolates obtained from St. Vincent’s Hospital (Sydney, Australia) and the University Hospital Galway (Galway, Ireland).The second approach used an extractionless method, whereby 30 known clinical isolates were obtained from University Hospital Galway and used for culture and broth samples.
Both approaches provided a sensitive and specific alternative to traditional non-molecular methods. Furthermore, two-thirds of clinical isolates tested had a mix of infections.
Table 3. Detection of ESBL and CPO resistant infections in QCMD samples using the EasyScreen™ ESBL/CPO Detection Kit, demonstrating identification of multiple targets in a single sample.
The accurate and rapid detection of these resistance genes using the EasyScreen™ ESBL/CPO Detection Kit supports diagnostic stewardship by providing diagnostic intelligence, readiness, and intervention, and informs antimicrobial stewardship programs. This ultimately supports improved health outcomes for patients, more appropriate treatment and management of antibiotic therapies, and offers significant cost savings.
Whilst Genetic Signatures offers a complete ESBL and CPO diagnostic solution via direct swab or via cultured methodology, the company also supports other diagnostic EasyScreen kits, automated instrumentation, service, and support. This allows hospitals and pathology laboratories to improve their operational efficiency and diagnostic workflow, ultimately promoting positive impacts to patient management and outcomes.
Learn more about Genetic Signatures products
Learn more about Genetic Signatures workflow solutions
Further information about Genetic Signatures
Genetic Signatures EasyScreen™ products are accessible in over 30 countries*, with direct representation in the US, United Kingdom, Germany, Australia, and distribution partners in Canada, Africa and other European nations. Products are used for IVD use and for research purposes. Please contact Genetic Signatures for more information, including the availability and use of the EasyScreen™ kits and workflow solutions in your region.
Please contact Genetic Signatures to determine the availability of EasyScreen™ products for IVD use in your region
References
- O’Neil, J. 2016, Tackling Drug-Resistant Infections Globally: Final Report and Recommendations, [Online], accessed 18/11/2021, <http://amr-review.org> (accessed 18/11/2021).
- OECD (2018), Stemming the Superbug Tide: Just A Few Dollars More, OECD Health Policy Studies, OECD Publishing, Paris, [online], accessed on 18/11/2021, <https://doi.org/10.1787/9789264307599-en>
- Martínez JL. 2008, Antibiotics and antibiotic resistance genes in natural environments. Science. 2008 Jul 18; 321(5887):365-7.
- National Museum of Australia (NMA), Penicillin, accessed on 17/11/2021, <https://www.nma.gov.au/defining-moments/resources/penicillin>
- OECD (2018), Stemming the Superbug Tide: Just A Few Dollars More, OECD Health Policy Studies, OECD Publishing, Paris, [online], accessed on 18/11/2021, <https://doi.org/10.1787/9789264307599-en>
- World Health Organisation (WHO), 2021, Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report: 2021, [online], accessed 17/11/2021, <https://www.who.int/publications/i/item/9789240027336>
- P. Magiorakos, A. Srinivasan, R. B. Carey, Y. Carmeli, M. E. Falagas, C. G. Giske, S. Harbarth, J. F. Hinndler et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria. Clinical Microbiology and Infection, Vol 8, Iss. 3 first published 27 July 2011 [via Wiley Online Library].
- OECD et al. 2017, Tackling Antimicrobial Resistance Ensuring Sustainable R&D, [online], accessed on 18/11/21, <http://www.oecd.org/els/health-systems/G20-AMR-Final-Paper-2017.pdf>
- National Database of Antibiotic Resistant Organisms (NDARO), [online], accessed on 18/11/2021, < https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/>
- UK Health Security Agency, 2021, English surveillance programme for antimicrobial utilisation and resistance (ESPAUR) Report 2020 to 2021, [online], accessed on 18/11/2021, <https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1033851/espaur-report-2020-to-2021-16-Nov.pdf>
- World Health Organisation (WHO), 2017, Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery and Development of New Antibiotics, [online], accessed on 18/11/2021, <https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf>
- Centre for Disease Control and Prevention (CDC), 2019, Biggest Threats and Data. [online], accessed on 18/11/2021, <https://www.cdc.gov/drugresistance/biggest-threats.html>
- Australian Commission on Safety and Quality in Health Care. National Alert System for Critical Antimicrobial Resistances (CARAlert), [online], accessed on 17/11/2021, https://www.safetyandquality.gov.au/our-work/antimicrobial-resistance/antimicrobial-use-and-resistance-australia-surveillance-system/national-alert-system-critical-antimicrobial-resistances-caralert
- Australian Commission on Safety and Quality in Health Care. AURA 2021: Fourth Australian report on antimicrobial use and resistance in human health. Sydney: ACSQHC; 2021. [online], Accessed on 18/11/2021. safetyandquality.gov.au/publications-andresources/aura-surveillance-system-reports-and-resources
- United Kingdom Government. 2021. AMRHAI reference unit: Reference and diagnostic services. [online], accessed on 18/11/2021. <https://www.gov.uk/guidance/amrhai-reference-unit-reference-and-diagnostic-services>
- American Society for Microbiology (AMR), 2021. Diagnostic Stewardship Interventions That Make a Difference. [Online] Accessed on 18/11/2021. <https://asm.org/Articles/2021/August/Diagnostic-Stewardship-Interventions-That-Make-a-D>
- Centre for Disease Control and Prevention (CDC). 2019. Core Elements of Hospital Antibiotic Stewardship Programs. [online], accessed on 18/11/2021. <https://www.cdc.gov/antibiotic-use/core-elements/hospital.html>
- Fang, F and Perez, K. 2020. University of Washington. Diagnostic Stewardship: A vision of the future of antimicrobial therapy. [online], Accessed on 18/11/2021. <https://www.cidrap.umn.edu/sites/default/files/public/downloads/diagnostic_stewardship_fang_and_perez_dec_17_2020.pdf>
- Maya Beganovic, Erin K McCreary, Monica V Mahoney, Brandon Dionne, Daniel A Green, Tristan T Timbrook, Interplay between Rapid Diagnostic Tests and Antimicrobial Stewardship Programs among Patients with Bloodstream and Other Severe Infections, The Journal of Applied Laboratory Medicine, Volume 3, Issue 4, 1 January 2019, Pages 601–616, https://doi.org/10.1373/jalm.2018.026450
- Dryden M, Johnson AP, Ashiru-Oredope D, Sharland M. Using antibiotics responsibly: right drug, right time, right dose, right duration. J Antimicrob Chemother. 2011 Nov;66(11):2441-3. doi: 10.1093/jac/dkr370. Epub 2011 Sep 15. PMID: 21926080.
- Messacar K, Parker SK, Todd JK, Dominguez SR. Implementation of Rapid Molecular Infectious Disease Diagnostics: the Role of Diagnostic and Antimicrobial Stewardship. J Clin Microbiol. 2017 Mar;55(3):715-723. doi: 10.1128/JCM.02264-16. Epub 2016 Dec 28. PMID: 28031432; PMCID: PMC5328439.
- World Health Organisation (WHO). 2016. Diagnostic stewardship. A guide to implementation in antimicrobial resistance surveillance sites. [online], Accessed on 18/11/2021. <https://apps.who.int/iris/bitstream/handle/10665/251553/WHO-DGO-AMR-2016.3-eng.pdf>
- Wolk, D. 2021. Geisinger Laboratories. (Cited by Prinzi, A. 2021. Diagnostic Stewardship Interventions That Make a Difference. American Society for Microbiology. [online], Accessed on 18/11/2021. https://asm.org/Articles/2021/August/Diagnostic-Stewardship-Interventions-That-Make-a-D)
- Lynette Phee, Stephanie Paget, Judy Jacques, Binutha Bharathan, Husam El-Mugamar, Anand Sivaramakrishnan, Carbapenemase-producing organism (CPO) colonisation at a district general hospital: universal screening may help reduce transmission, Infection Prevention in Practice, Volume 3, Issue 3, 2021, 100164, ISSN 2590-0889, https://doi.org/10.1016/j.infpip.2021.100164.
- Gonzalez, C.; Oueslati, S.; Biez, L.; Dortet, L.; Naas, T. Evaluation of the EasyScreen™ ESBL/CPO Detection Kit for the Detection of ß-Lactam Resistance Genes. Diagnostics 2022, 12, 2223
- Hoyos-Mallecot, Y.; Ouzani, S.; Dortet, L.; Fortineau, N.; Naas, T. Performance of the Xpert Carba-R v2 in the daily workflow of a hygiene unit in a country with a low prevalence of carbapenemase-producing Enterobacteriaceae. Int. J. Antimicrob. Agents 2017, 49, 774–777