Our Technology
3base® Technology
Unique solutions for syndromic testing for infectious disease
Simplify to Amplify
Our scientists are pioneers of the bisulphite method used for distinguishing methylated cytosine (Methyl C) residues from unmethylated cytosine (C) residues in DNA.
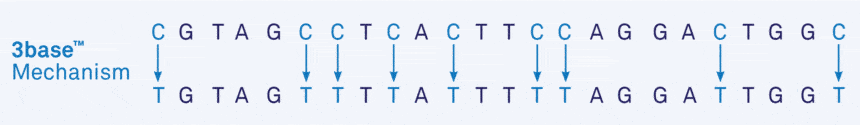
The technology not only serves to identify methylation patterns in the DNA, it can also be used to simplify DNA and RNA sequences more generally, particularly in microorganisms. In unmethylated nucleic acid, the bisulphite method converts all cytosines (C) ultimately into thymine (T), so that Cs disappear from the sequence altogether, resulting in the conversion of a 4 base sequence into a 3 base sequence of only As, Ts and Gs.
We refer to this resultant DNA or RNA as ‘3base®’.
Advantages of 3base® technology
Increased sequence homology
By eliminating one-in-four of the naturally occurring bases, 3base® technology is unique in its ability to simplify the detection of an entire class of microbial species. This elimination of many sub-species variations is a key intrinsic advantage of Genetic Signatures’ 3base® technology.
- Figure 1a. Example of the 3base® mechanism. The example sequences below show the increase in homology from 75% (“Before”) to 95% (“After”) via the 3base® conversion where all C bases are detected as T bases.
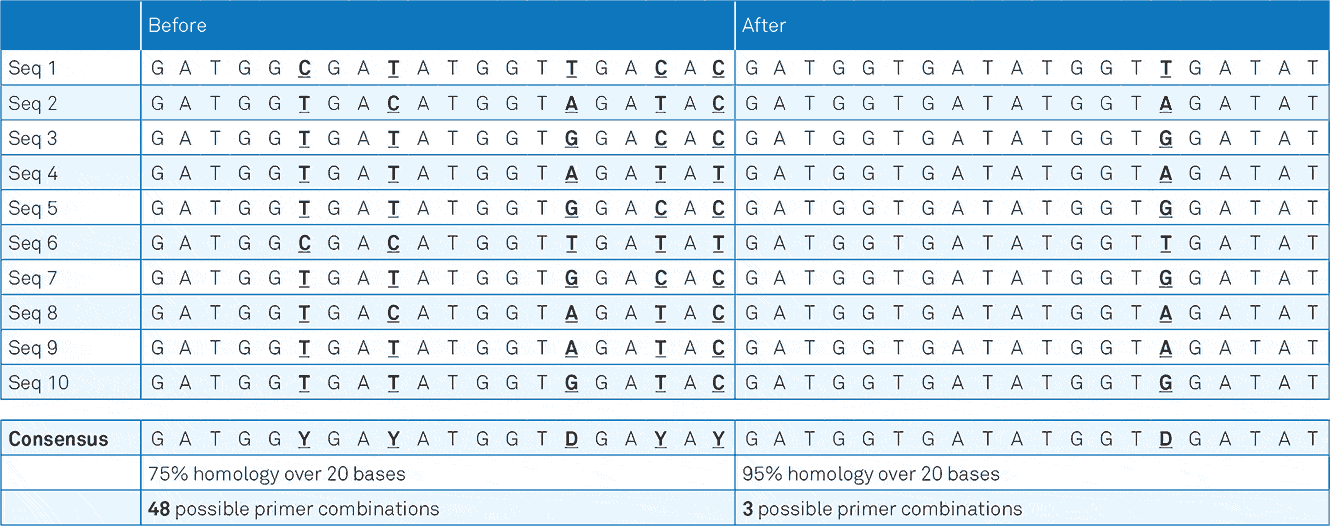
The 3base® converted sequence, now only contains 1 variable position requiring only 1 degeneracy, or mixed-base in the primer. This increases the homology from 75% to 95% and decreases the number of different primer species from 48, to just 3.
This simplifies syndromic multiplex real-time PCR detection as 3base®;
- Significantly improves consensus primer homology across species variants
- Enables the use of single or minimal primers/probes to accurately screen for all/any variants present in a sample
Simplified multiplex PCR
Efficient pan-screening for a wide range of infectious diseases
- 3base® increases the sequence similarity to expand screening for pathogen subtypes, without compromising on specificity or sensitivity
- Hence, the technology is more 'inclusive' to allow subtype / variant detection of a target pathogen
Reduction of G-C regions for efficient PCR amplification
3base® removal of C-nucleotides supports more uniform PCR primer melting temperatures, thereby improving PCR efficiency, such as reducing non-specific binding. This allows more harmonised amplificant conditions to support efficient multiplex PCR due to;
- more efficient multiplex assays
- fewer reagents required with more harmonised reaction conditions
More efficient multiplex PCR assays
- Fewer primers, less competition and expanded screening of pathogen families without compromising assay sensitivity or specificity
Greater ‘resilience’ to genetic variation
- 3base® improves resilience to both C→T and T→C mutations, the most frequently occurring mutation in viral genomes
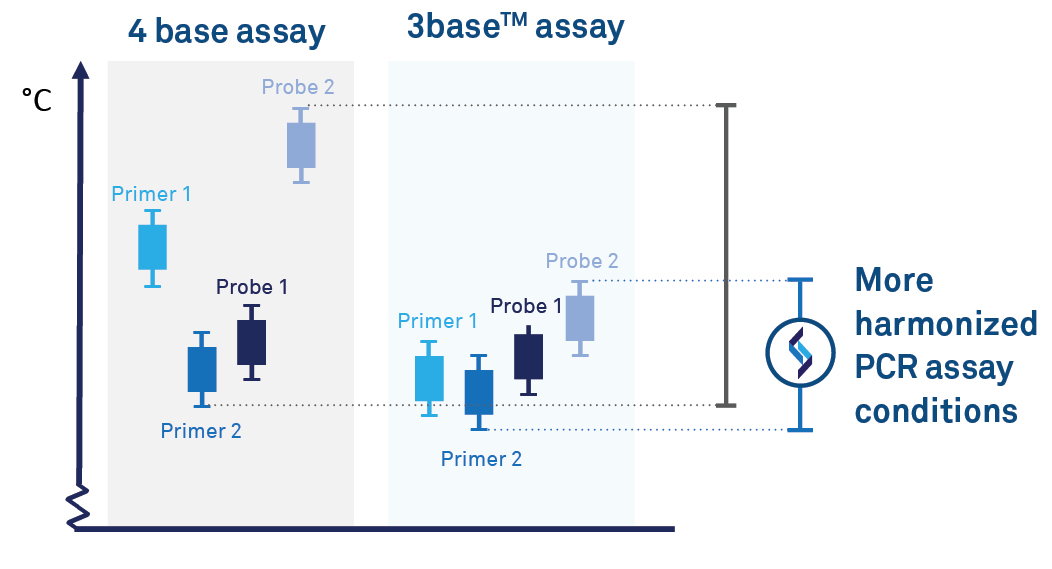
Conversion of C-T within target sequences harmonizes primer and
probe sets. This can improve PCR efficiency, particularly for detecting
organisms with G-C rich genomes.
Unique sample processing method is more efficient at lysing difficult pathogens such a parasites
For example, parasites such as microsporidia.
Listen to the recent webinar on the Advances in Molecular Detection of Gastrointestinal Parasites here
Read the recent scientific publication demonstrating the power of 3base® technology here:
Diagnosis of Viral Families Using Nucleic Acid Simplification Technique
By Dr. Douglas Millar and Dr. John Melki
