Omicron BA.4 and BA.5 Subvariants
Contents
- Epidemiology
- Healthcare Burden and Public Health Response
- 3base™ Technology
- Confident Detection of All Known SARS-CoV-2 Variants with 3base™
- Customer Experience with the EasyScreen™ SARS-CoV-2 Detection Kit (RP012)
- Contact your local team
- References
Summary
- Omicron BA.4 and BA.5 subvariants now represent over half of all COVID-19 infections globally, due to new mutations enhancing viral transmission and immune evasion.
- Health authorities are encouraging vaccination and additional boosters to support immunity and reduce the risk of severe infection, with health authorities reducing the reinfection period from 12 weeks to just 28 days.
- PCR remains the gold-standard for the molecular detection of SARS-CoV-2, and can be integrated with broader syndromic testing for the detection of other respiratory pathogens that cause similar signs and symptoms.
- Genetic Signatures 3baseTM technology enables confident detection of all known SARS-CoV-2 variants due to improved ‘immunity’ to genetic mutations.
Contact your local Genetic Signatures representative for more information here.
Epidemiology
The recently identified SARS-CoV-2 Omicron BA.4 and BA.5 subvariants have rapidly become dominant strains in many countries. These novel subvariants originated from South Africa in January and February 2022 respectively and have quickly spread across the globe due to additional mutations improving transmission, and enhancing the virus’ ability to evade the immune system of previously infected and vaccinated populations.1,2 However, vaccine immunity continues to provide substantial protection against severe disease with Omicron BA.4 and BA.5 and hence, health authorities are encouraging additional booster vaccinations.
The World Health Organization (WHO) designated Omicron as a Variant of Concern (VOC) in November 2021 (read more in our blog article here), with BA.4 and BA.5 subvariants immediately placed ‘under monitoring’ since detection in January 2022.3 Omicron BA.4 and BA.5 are noted as substantially (4.2-fold) more resistant to sera from vaccinated and boosted individuals from the original SARS-CoV-2 virus and Omicron BA.2.12.1 variant, and therefore, are more likely to contribute to future outbreaks. 3
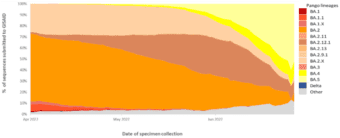
Indeed, Omicron BA.4 and BA.5 are now the dominant strains in Africa and are predicted to dominate European infections by July 2022.4,5 Omicron BA.4 and BA.5 have also seen rapidly increased infection rates in the United States, estimated to represent 16% and 65% respectively, of new COVID-19 infections in early July 2022.6 These rates also align with reported cases in Australia, with the exception of BA.2, which is recorded at a higher rate than the US.7 A global overview of the VOC infection rates is shown in Figure 1, sourced from the World Health Organization’s weekly COVID-19 update to 3 July 2022.8 This data shows that Omicron BA.5 jumped from 14% to represent over half (51.88%) of all COVID-19 infections globally in just four weeks, as of the last week of June, 2022. 8
Figure 1. Global VOC rates indicate surge in BA.4 and BA.5 subvariants
This graph shows the percentage of all circulating variants since 1 April 2022. Omicron sister-lineages and additional Omicron VOC descendent lineages under further monitoring (VOC-VUM) are shown. BA.1.X and BA.2.X include all BA.1 and BA.2 pooled descendent lineages, except those already shown in the figure above. Source: SARS-CoV-2 sequence data and metadata from GISAID, as of 3 July 2022. 8
Healthcare Burden and Public Health Response
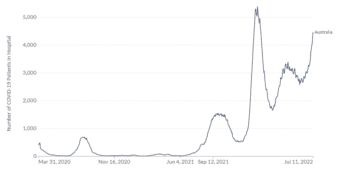
Australia’s experience of COVID-19 and other non-SARS-CoV-2 respiratory pathogens carries important lessons for the northern hemisphere, yet to enter the 2022 winter season. 9,10 A sharp increase in COVID-19 associated hospitalisations has also been seen in Australia, reaching 4468 hospitalisations on July 11th compared to 2682 the week prior, shown in Figure 2.11 In addition, data from the Omicron wave during the period 15 Dec 2021 – 5 June 2022 indicated 5902 deaths in Australia due to COVID-19.12,13 Whilst the spread of COVID-19 variants is a significant health threat, the testing and surveillance of other respiratory pathogens globally remains important, with higher infection rates for many pathogens in 2022 compared to those seen during 2020 and 2021, following relaxing of social distancing and travel restrictions.7,14
Figure 2. COVID-19 hospitalisations in Australia 27 March 2020 to 11 July 2022
Official data collated by Our World in Data11 – Last updated 12 July 2022
Health professionals are concerned about the increased rate of Omicron BA.4/5 reinfection, with Australia reducing the time period of reinfection from 12 weeks to just 28 days.15 The Australian Health Protection Principal Committee stated that positive tests detected after 28 days from an original infection will be counted as new cases, and mask wearing has been strongly encouraged in all indoor spaces, public transport and aged care facilities. This will also help reduce the spread of other respiratory illnesses frequently seen during winter months.15
3base™ Technology
Whilst rapid antigen testing has supported self-testing to detect SARS-CoV-2 infection, PCR testing remains the gold-standard due to improved sensitivity and accuracy of detection. Health experts also advise that RATs cannot achieve the same level of surveillance as PCR testing approaches, which can provide a false sense of security from negative test results, with reports by the FDA of the new Omicron variants yielding a true RAT result in only 60% of positive cases.16,17 In addition, compared to rapid antigen approaches, the alignment of PCR assays can be compared with genomes of new viral strains over time to ensure tests maintain performance. Unlike traditional 4 base PCR assays, Genetic Signatures unique 3baseTM technology provides ‘immunity’ to the vast majority of the genetic variation that occurs in SARS-CoV-2 variants.18 The 3baseTM process converts the naturally occurring 4-base genome to one of just 3 bases, due to the conversion of Cytosine (C) to Thymine (T), shown in Figure 3. This significantly reduces sequence variation in genomes. The improved sequence similarity between subtypes of related pathogens supports the effectiveness of screening for known subtypes or gene families, whilst also making 3base™ assays more ‘immune’ to random sequence variation in target organisms, which may affect the efficiency of conventional 4-base assays.19,20
Figure 3. Genetic Signatures 3base™ mechanism, where cytosines are converted into thymine.
Cytosine is the nucleotide base that mutates most frequently. The frequency of this spontaneous mutation is 140-fold higher in ssRNA molecules (such as SARS-CoV-2) compared to double stranded molecules. 21 Genetic Signatures in silico analysis has confirmed the high percentage of mutations in the BA.4 and BA.4 variants being of a C>T or T>C mismatch, which don’t affect the target sequence Genetic Signatures’ assays are designed to detect. This is discussed in more detail in the next section.
Advantages of 3base™ technology include:
- Immunity to genetic variation: 3base™conversion of C to T also provides ‘immunity’ to potential genetic variation that may arise at random in organisms and was demonstrated with Genetic Signatures’ SARS-CoV-2 assay where 3baseTM enabled immunity to both C>T and T>C mutations, with C>T being the most frequently occurring spontaneous mutation in nucleic acids21,22
- Detection of subtypes: Due to increased sequence similarity, fewer primers can be used for the detection of known subtypes or gene families, providing screening capabilities for a wide number of clinically relevant targets19
- More efficient multiplex assays: Fewer primers, less competition and improved sensitivity and specificity
- Reduced G-C content: Conversion of C to T bases within target sequences contributes to improved PCR efficiency for organisms with high G-C content such as Giardia, where inefficiencies with amplification have been reported using 4 base assays20
- Uniform sample processing conditions: 3base™provides a universal extraction process regardless of sample type, and is compatible with both DNA and RNA from bacteria, protozoa, viruses and fungi
- Reduced contamination: 3base™ conversion of samples ensures 3base™ assays are not affected by environmental contamination by native 4 base sequences
Learn more about Genetic Signatures’ unique 3base technology here
Confident Detection of All Known SARS-CoV-2 Variants with 3base™
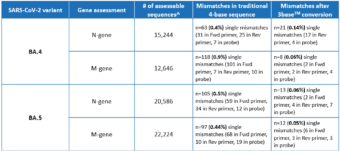
A recent in silico analysis by Genetic Signatures of the Omicron BA.4 and BA.5 genome, derived from sequences submitted to the GISAID database between June 1st – June 28th 2022, assessed the alignment of the EasyScreen™ SARS-CoV-2 Detection Kit (RP012) to the N-gene and M-gene targets. Results in Table 1 show that 99.9% of reported sequences had 100% alignment to the assay, and demonstrated that 3base™ conversion of the 4-base sequence eliminated mismatches in the target regions, improving confidence in detection for the BA.4 and BA.5 variants. The data also demonstrates that mismatches of the BA.5 N-gene comprised 87% C and T mutations, which were rendered redundant by the 3baseTM conversion process.
Genetic Signatures has previously shown that the limit of detection (LOD) of the EasyScreen™ SARS-CoV-2 Detection Kit (RP012) is maintained at the same sensitivity as the original reference SARS-CoV-2 virus, confirmed with Alpha, Beta, Delta, Gamma, Lambda and Omicron (1.1.529) culture.
Table 1. Global In Silico Analysis performed June 2022 for the BA.4 and BA.5 Subvariants
^Assessable sequences have 100% coverage of RP012 target regions and no ambiguous bases
Learn more about our Respiratory product range here
The EasyScreen™ SARS-CoV-2 Detection Kit has TGA and CE-IVD registration, and is supplied in the US under the FDA Section IV.c exemption of the Policy for Coronavirus Disease-2019 Tests During the Public Health Emergency (Revised).
The EasyScreen™ Sample Processing Kit (SP012) has TGA and CE-IVD registration, and is supplied in the US as FDA-listed clinical concentrators but not for in vitro diagnostic use.
The EasyScreen™ SARS-CoV-2 Variant Detection Kit is for research use only and not for in vitro diagnostic use.
Customer Experience with the EasyScreen™ SARS-CoV-2 Detection Kit (RP012)
KH Labor GmbH, a provider of laboratory diagnostic services across multiple sites in Germany, has been a valued Genetic Signatures’ customer since the end of 2020. Genetic Signatures has developed a close partnership with Dr. Mark Wasner, Technical Head of Microbiology and Molecular Diagnostics, and his laboratory team, meeting the high throughput SARS-CoV-2 PCR testing requirements throughout the COVID-19 pandemic. The EasyScreen™ SARS-CoV-2 Detection Kit is run on Genetic Signatures GS-1000 automated sample processing system, providing flexibility to scale up or down, depending on the testing requirements.
The advantages of Genetic Signatures’ 3base™ technology gave KH Labor confidence in detecting known and new SARS-CoV-2 variants. In addition, their laboratory workflow was further improved by the recent implementation of Genetic Signatures’ SARS-CoV-2 PCR FAST protocol, which significantly reduces ‘time to result’ and improves throughput due to faster thermocycling.
References:
- Epidemiological update: SARS-CoV-2 Omicron sub-lineages BA.4 and BA.5, [online], accessed 13/7/22, https://www.ecdc.europa.eu/en/news-events/epidemiological-update-sars-cov-2-omicron-sub-lineages-ba4-and-ba5
- Wang, Q., Guo, Y., Iketani, S. et al. (2022), Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4, & BA.5. Nature, https://doi.org/10.1038/s41586-022-05053-w
- Tracking SARS-CoV-2 Variants, [online], accessed 13/7/22, https://www.who.int/activities/tracking-SARS-CoV-2-variants
- Implications of the emergence and spread of the SARS-CoV-2 variants of concern BA.4 and BA.5 for the EU/EEA, [online], accessed 13/7/22, https://www.ecdc.europa.eu/en/news-events/implications-emergence-spread-sars-cov-2-variants-concern-ba4-and-ba5
- New coronavirus subvariants escape antibodies from vaccination and prior Omicron infection, studies suggest, [online], accessed 13/7/22, https://edition.cnn.com/2022/06/22/health/ba4-ba5-escape-antibodies-covid-vaccine/index.html
- COVID Data Tracker, [online], accessed 13/7/22, https://covid.cdc.gov/covid-data-tracker/#variant-proportions
- NSW Respiratory Surveillance Report – week ending 02 July 2022, [online], accessed 13/7/22, https://www.health.nsw.gov.au/Infectious/covid-19/Documents/weekly-covid-overview-20220702.pdf
- Weekly epidemiological update on COVID-19 – 6 July 2022, [online], accessed 13/7/22, https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19—6-july-2022
- Hodjat, P., Christensen, P. A., Subedi, S., Bernard, D. W., Olsen, R. J., & Long, S. W. (2021), The reemergence of seasonal respiratory viruses in Houston, Texas, after Relaxing COVID-19 restrictions, Microbiology Spectrum, 9(2)
- Olsen SJ, Winn AK, Budd AP, et al. (2021), Changes in Influenza and Other Respiratory Virus Activity During the COVID-19 Pandemic — United States, 2020–2021. MMWR Morb Mortal Wkly Rep, 70:1013–1019
- Our World in Data, number of COVID-19 patient in hospital, [online], access 13/7/22, https://ourworldindata.org/grapher/current-covid-patients-hospital?country=AUS
- COVID-19 Australia: Epidemiology Report 62 COVID-19 National Incident Room Surveillance Team Reporting period ending 5 June 2022, Communicable Diseases Intelligence 2022 , Volume 46, [online], accessed 13/7/22, https://www1.health.gov.au/internet/main/publishing.nsf/Content/C50CAE02452A48A7CA2587320081F7BF/$File/covid_19_australia_epidemiology_report_62_reporting_period_ending_5_june_2022.pdf
- Provisional Mortality Statistics, [online], accessed 13/7/22, https://www.abs.gov.au/statistics/health/causes-death/provisional-mortality-statistics/latest-release
- FLUVIEW interactive, National, Regional and State Level Outpatient Illness and Viral Surveillance, [online], accessed 13/7/22, https://gis.cdc.gov/grasp/fluview/fluportaldashboard.html
- Immune period for COVID positive cases could be reduced from 12 weeks to 28 days, [online], accessed 13/7/22, https://www.abc.net.au/news/2022-07-11/sa-immune-period-for-covid-positive-cases-reduced-from-12-weeks-/101226156
- COVID-19 antigen test sensitivity could be as low as 60% with omicron: FDA, [online], accessed 18/7/22, https://www.medtechdive.com/news/covid-antigen-test-sensitivity-low-omicron/626461/
- Friday Q&A: Ian Lipkin, Columbia epidemiologist, talks COVID-19 testing as U.S. sees another wave, [online], accessed 18/7/22, https://www.medtechdive.com/news/columbia-epidemiologist-COVID-variant-testing/627306/?:%202022-07-15%20MedTech%20Dive%20%5Bissue:43131%5D
- Frommer M et al. (1992), A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands, Proc Natl Acad Sci U S A, 89 (5): 1827–1831
- Starr K, et al. (2019), Comparison of Three Adenovirus Quantitative PCR Assays with ATCC Reference Strains and Clinical Samples. J Clin Microbiol., 57(11):e00735-19
- Rochelle PA, et al. (1997), Comparison of primers and optimization of PCR conditions for detection of Cryptosporidium parvum and Giardia lamblia in water. , Appl Environ Microbiol., 63:106–14.
- Genetic Signatures, 11 Oct 2021, Validation of the EasyScreen 3base SARS-CoV-2 Detection Kit RP012 on Saliva Samples. NRL Workshop – 11-13th of October 2021, [online], accessed on 14/12/2021, https://geneticsignatures.com/au/resource/nrl-presentation-2021/
- Frederico LA et al., (1990), A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy, Biochemistry, (10):2532-7