3base Technology and Reliable Detection of SARS-CoV-2 Variants
Genetic Signatures announces the launch of their multiplex real-time SARS-CoV-2 Variant Detection Kit, which can specifically identify Omicron (B.1.1.529) and other key SARS-CoV-2 variants.
Summary
Genetic Signatures announces the development of their SARS-CoV-2 Variant Detection Kit, which can specifically identify the Omicron (B.1.1.529), Delta and Beta variants by detecting specific mutations, and includes a positive control target for SARS-CoV-2. This complements the company’s existing kit, the SARS-CoV-2 Detection Kit, which is able to effectively screen for all known variants by identifying two gene targets common to all strains.
The newly developed multiplex real-time PCR assay for variant detection will assist laboratories to understand Omicron’s epidemiology and incidence in the community, and support the prioritisation of suspected Omicron positive patient samples for genomic sequencing.
Genetic Signatures’ patented 3base™ technology provides a fundamental advantage for addressing the highly evolving SARS-CoV-2, with the unique technology more ‘immune’ to mutations than traditional 4-base PCR diagnostic techniques.
Despite 60 mutations detected in the Omicron variant compared to the original Wuhan variant, Genetic Signatures’ existing EasyScreen™ SARS-CoV-2 Detection Kit identifies all variants tested with the same limit of detection as the original reference SARS-CoV-2 virus, providing laboratories confidence in detecting the rapidly evolving SARS-CoV-2.
Genetic Signatures’ rapid response to produce the EasyScreen™ SARS-CoV-2 Variant Detection Kit showcases the world class expertise of the Research and Development team.
Contact your local Genetic Signatures representative for more information here.
Contents
- Molecular detection of SARS-CoV-2
- SARS-CoV-2 variant evolution and its implications
- B.1.1.529 Omicron variant
- Mutational molecular immunity: 3base technology
- EasyScreen™ SARS-CoV-2 Detection Kit
- Molecular identification of the B.1.1.529 Omicron variant
- EasyScreen™ SARS-CoV-2 Variant Detection Kit
- References
Molecular detection of SARS-CoV-2
Since the SARS-CoV-2 virus was first identified in Wuhan, China in December 2019, the world quickly came to know this virus as the causal agent of Coronavirus Disease 2019 (COVID-19). Despite significant efforts to contain infection, SARS-CoV-2 quickly spread across the globe, ultimately leading the World Health Organization (WHO) to declare a pandemic just 3 months later, in March 2020.
Due to significant COVID-19 morbidity and mortality, early detection and identification of SARS-CoV-2 became a key line of defence, assisting with surveillance, management and containment, and early treatment. Precise and rapid molecular diagnostic assays, enabled by real-time reverse transcription-polymerase chain reaction (RT-PCR), quickly became the gold standard for detecting SARS-CoV-2 over other respiratory infections, with high sensitivity and specificity, and also in high throughput [1, 2, 3].
The molecular detection of SARS-CoV-2 involves the use of specific primers and probes that match the viral genetic sequence, to amplify a small segment of the targeted viral genetic material using real-time PCR. Viral RNA is extracted from the clinical specimen and purified RNA is reverse transcribed into complementary DNA (cDNA). The cDNA subsequently undergoes amplification using PCR to amplify the gene of interest, monitored in real-time by detection of fluorescence.
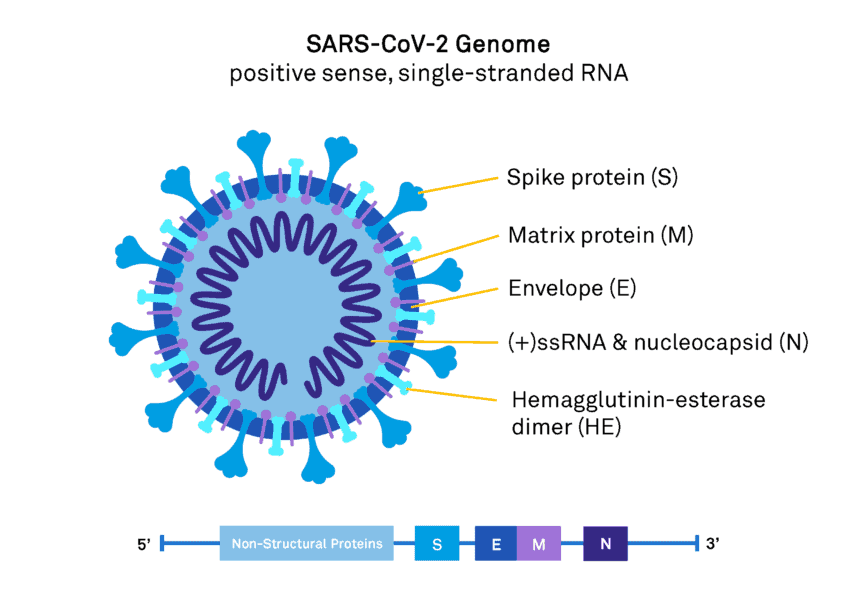
Real-time PCR primers target the more highly conserved and abundantly expressed genes of SARS-CoV-2 including the structural spike proteins (S), nucleocapsid (N), matrix protein (M), non-structural RNA-dependent RNA polymerase (RdRp), and the open reading frame ORF1ab (Figure 1). These genes have higher sensitivity for coronavirus detection [4, 5].
Figure 1. SARS-CoV-2 genome and viral structure.
PCR is a highly sensitive and specific tool for detecting genetic material of extremely small amounts. PCR tests for SARS-CoV-2 are generally designed to identify multiple gene targets to minimise the chance of false-negative results occurring due to possible mutations in the target regions. However, test sensitivity may occasionally be impacted if a mutation occurs in the part of the genome specifically assessed by that test [6, 7, 8]. Positive samples identified by PCR are also able to be sent for DNA sequencing to confirm the result providing an opportunity to sequence the viral genome for the surveillance of variants.
SARS-CoV-2 variant evolution and its implications
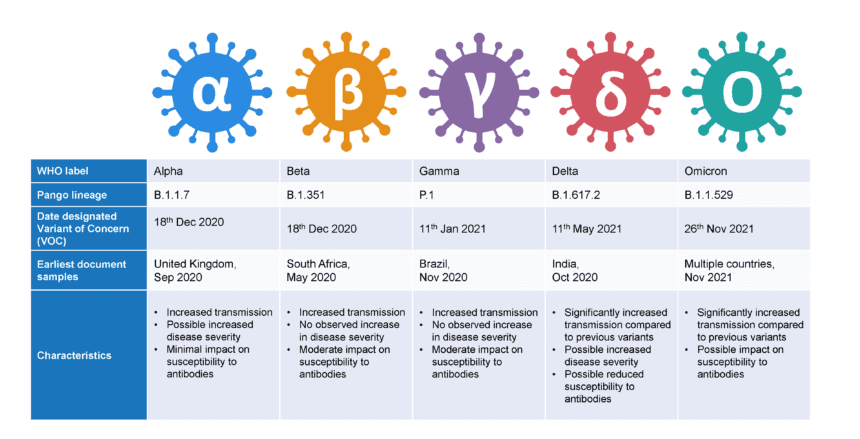
Indeed, SARS-CoV-2 is a rapidly evolving virus. Since the original “L” type virus was sequenced, many other variants have arisen with mutations in the spike protein and to a lesser extent, the nucleocapsid protein and non-structural proteins, potentially increasing the virus’ ability to spread and cause disease. The emergence of variants that posed an increased risk to global public health prompted the characterisation of specific Variants of Interest (VOIs) and Variants of Concern (VOCs) [9]. To date, over 6,000 variants have been documented in the publicly available GISAID database, which houses over 6 million SARS-CoV-2 genome submissions [10]. Whilst most variants have emerged and then disappeared, others have persisted and caused higher transmission rates and / or disease. Currently 2 variants are designated by the WHO as VOI and 5 as VOC. VOC include the Alpha, Beta, Gamma, and Delta SARS-CoV-2 variants, with the recently detected B.1.1.529 Omicron variant identified in November 2021 (Figure 2) [9].
Figure 2. World Health Organisation (WHO) Variants of Concern (VOCs) (9, 11).
B.1.1.529 Omicron variant
The B.1.1.529 variant was first detected in Botswana, South Africa, on November 11, 2021. By the 24th of November, Omicron was reported by the World Health Organisation (WHO) and two days later, designated as a VOC [11). In just over a month, Omicron had already been detected in almost 60 other countries around the world, many reporting locally acquired cases [10, 12, 13].
Some 60 mutations have been identified in the Omicron variant genome, compared to the original Wuhan SARS-CoV-2 virus, with 32 changes to the spike protein gene (S-gene), including the receptor binding domain and furin cleavage sites, potentially changing the behaviour of the virus. In addition, a range of mutations were also identified in typically conserved genes including within ORF1ab, the N-gene, and the M-gene [14].
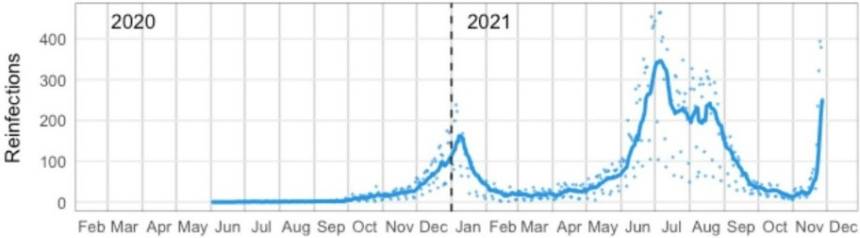
Scientists around the globe are working to determine how these mutations, particularly in the spike protein, will impact transmission, infection, the effectiveness of the existing vaccines, and natural antibodies. Initial studies suggest that infection-induced and vaccination-induced immunity is not as effective at blocking Omicron from entering cells (where neutralizing antibodies play a significant role), explaining higher re-infection rates and detection amongst vaccinated populations (Figure 3) [15, 16, 17]. Indeed, reported PCR results from the UK indicate that high levels of SARS-CoV-2 immunity is not protective against Omicron infection [18].
Figure 3. Suspected daily numbers of SARS-CoV-2 reinfection in South Africa [15, 16]. The solid blue line indicated the 7-day moving average. Blue points are daily values (Imaged sourced from Pulliam et al, 2021 Preprint).
However, B-cell and T-cell immunity induced by authorized vaccines is expected to remain effective against severe illness, hospitalizations, and deaths, and assist in reducing the risk of long COVID [19]. Therefore, the importance of vaccination and booster vaccinations remains. Further research is required to determine how effective antibody protection against the Omicron variant will be and thus, the importance of SARS-CoV-2 variant detection and identification is essential for understanding the epidemiology of this virus.
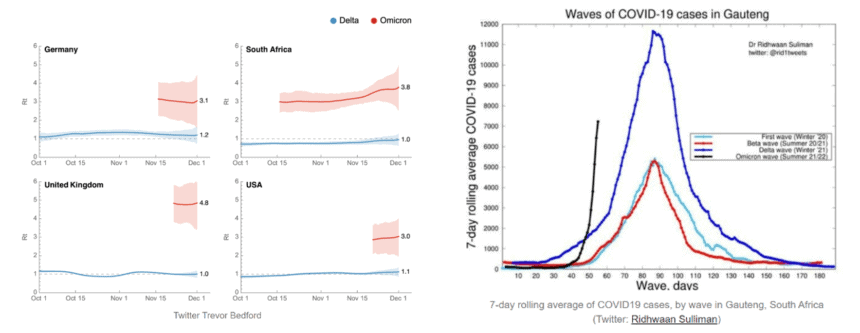
Omicron appears to be spreading faster than many countries can detect it – and at a rate that has not been seen for other SARS-CoV-2 variants (Figure 4) [18, 20]. Rapid molecular SARS-CoV-2 detection remains the gold standard for detecting Omicron. However, some molecular diagnostic SARS-CoV-2 kits targeting the mutated S-gene sequences have potentially resulted in S-gene target failure (SGTF). It has been suggested that laboratories using these molecular PCR detection kits targeting the S-gene will likely underrepresent the volume of Omicron cases due to false negative reporting (21). This presents additional health and societal challenges for early patient treatment, containing community spread and contact tracing.
The United States (US) Food and Drug Administration (FDA) has recently reported 27 assays from 23 developers and manufacturers “whose performance may be impacted by mutations in the SARS-CoV-2 Omicron variant.” In addition, a test detecting a single region in the mutated Omicron N-gene was also listed, of which 4 nucleotide changes were identified [10, 14]. Impacted assays are listed here [8].
Figure 4. 4a) SARS-CoV-2 estimated reproduction rate (Rt) (Image sourced from Trever Bedford, cited by Jetelina, 2021 [20]). 4b) COVID-19 cases in South Africa (Image sourced from Dr Ridwaan Silliman, cited by Jetelina, 2021 [18]).
The importance of mutational immunity of molecular diagnostic screening to detect Omicron and future SARS-CoV-2 variants is clear. 3base™ technology meets this challenge, providing reliable molecular detection of mRNA viruses, including SARS-CoV-2, providing laboratories with confidence in their reporting of SARS-CoV-2.
Mutational molecular immunity: 3base™ technology
3base™ technology, unique to Genetic Signatures, significantly reduces sequence variation in genomes to improve similarity between subtypes of related pathogens, effectively making 3base™ assays more ‘immune’ to sequence variation, which may affect the efficiency of conventional 4-base assays.
Cytosine is the nucleotide base that mutates most frequently. The frequency of this spontaneous mutation is 140-fold higher in ssRNA molecules (such as SARS-CoV-2) compared to double stranded molecules (22).
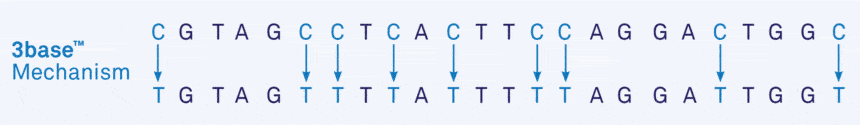
The 3base™ process converts all cytosines (C) into thymine (T), so that Cs disappear from the sequence altogether, resulting in the conversion of a 4-base sequence into a 3-base sequence of only As, Ts and Gs, referred to as ‘3base™’ (Figure 5).
Figure 5. Genetic Signatures 3base™ mechanism, where unmethylated cytosines are converted into thymine.
Advantages of 3base™ technology include:
- Increased sequence similarity: Enables detection of sub-species and possible variants present in a sample exhibiting ‘immunity’ to genetic variation that may occur.
- More efficient multiplex assays: Fewer primers, less competition, improved sensitivity.
- Uniform assay conditions: Uniform temperature conditions for different primer and probe sets enables faster thermocycling times and universal application to all specimen types.
- Reduced cross reactivity: Particularly important when multiple strains of the same organism are present.
- Open platform: Compatible with most automated sample preparation systems and PCR instrumentation.
For more information on 3base™ technology, click here.
In silico analysis of the EasyScreen™ SARS-CoV-2 Detection Kit
Genetic Signatures performs in silico analysis of the EasyScreen™ SARS-CoV-2 Detection Kit. Of the ‘available sequences’ in the GISAID database, the ‘assessable sequences’ (with 100% coverage of the RP012 target regions) are used to identify the number of reported sequences with 100% alignment to the target regions.
In July 2021, Genetic Signatures’ EasyScreen™ SARS-CoV-2 Detection Kit was assessed against 290,863 SARS-CoV-2 M-gene sequences in the GISAID database. Of these sequences, 98% were a 100% match to the 4-base sequence. Of the 4,602 mismatched sequences, 81% were due to C/T or T/C mutations and thus, are rendered ‘invisible’ with 3base™ conversion, providing assay immunity to SARS-CoV-2 genetic drift, compared to a traditional 4-base assay which would have seen reduced assay efficiency and misaligned primers or probes [22].
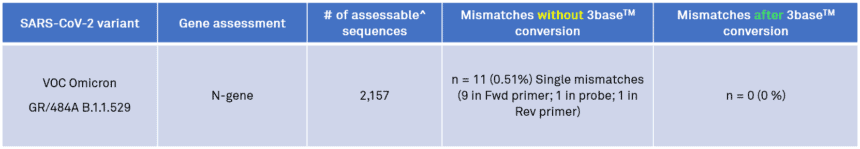
On the 12th of December 2021, 2409 sequences in the Omicron GR/484A/B.1.1.529 clade were analysed by in silico analysis for alignment to the Genetic Signatures’ EasyScreen™ SARS-CoV-2 Detection Kit. Again, the 3base™ conversion of the 4-base Omicron sequence eliminated all mismatches in the N-gene target region, thereby maintaining 100% alignment to the assessable sequences, further demonstrating improved immunity of 3base™ assays to genetic mutation (Table 1).
Table 1. In silico analysis of the EasyScreen™ Detection Kit RP012 to 2,157 sequences in the Omicron GR/484A/B.1.1.529 clade demonstrates 3baseTM conversion effectively removes all nucleotide mismatches in the N-gene target region to enable efficient PCR detection of the B.1.1.529 Omicron variant in clinical samples.
^Assessable sequences have 100% coverage of RP012 target regions and no ambiguous bases
Molecular identification of the B.1.1.529 Omicron variant
In November 2021, in response to the B.1.1.529 Omicron variant, the WHO asked the international community to (11):
- Enhance surveillance and sequencing efforts
- Submit complete genome sequences to a publicly available database, such as GISAID.
- Report initial cases/clusters associated with VOC infection to WHO.
- Perform field investigations and laboratory assessments to improve understanding of the COVID-19 epidemiology, severity, effectiveness of public health and social measures, diagnostic methods, immune responses, antibody neutralization, etc.
From a diagnostic perspective, the requirement to detect and sequence every positive SARS-CoV-2 to confirm every Omicron variant has placed enormous pressure on sequencing laboratories.
In response, Genetic Signatures has developed a SARS-CoV-2 Variant Detection Kit to assist with the detection of specific mutations of the SARS-CoV-2 variants in patient samples confirmed to contain SARS-CoV-2. The kit provides preliminary identification of SARS-CoV-2 Omicron and Delta variants, supporting the prioritisation of suspected Omicron positive samples for genomic sequencing, and providing research and clinical laboratories additional tools for tracking, and understanding the variant’s epidemiology.
EasyScreen™ SARS-CoV-2 Variant Detection Kit
The newly developed EasyScreen™ SARS-CoV-2 Variant Detection Kit uses 3base™ multiplexed Real-Time PCR assays, employing primer and probe sets specific for the variant mutations of interest and can be run in conjunction, or as a reflex, with the existing EasyScreen™ SARS-CoV-2 Detection Kit, which can detect, but not differentiate, all reported SARS-CoV-2 sequences.
The EasyScreen™ SARS-CoV-2 Variant Detection Kit is available for manual or high throughput assays, supported by the automated workflow on the GS1-HT or GS-1000 instruments, with fast assay capabilities. The assay is compatible with swabs (dry, or in liquid media) samples.
The new EasyScreen™ SARS-CoV-2 Variant Detection Kit was specifically designed in collaboration with customers to detect and identify the Omicron and Delta variant via their specific mutations prior to sequencing, and has been confirmed against more than 200 clinical samples. The test is intended to be used by testing laboratories, either in parallel with the primary screen, or as a reflex test performed once positive cases have been identified. It is one of only a few tests globally that can positively identify a key Omicron mutation prior to sequencing.
The rapid response to produce the SARS-CoV-2 Variant Detection Kit following the WHO designating Omicron as a VOC showcases Genetic Signatures’ innovative culture and world class expertise of the Research and Development team.
For availability and regulatory approval of the EasyScreen SARS-CoV-2 Detection Kit and EasyScreen SARS-CoV-2 Variant Detection Kit in your region, contact your local Genetic Signatures representative here.
The EasyScreen™ SARS-CoV-2 Detection Kit has TGA and CE-IVD registration, and is supplied in the US under the FDA Section IV.c exemption of the Policy for Coronavirus Disease-2019 Tests During the Public Health Emergency (Revised).
The EasyScreen™ Sample Processing Kit (SP012) has TGA and CE-IVD registration, and is supplied in the US as FDA-listed clinical concentrators but not for in vitro diagnostic use.
The EasyScreen™ SARS-CoV-2 Variant Detection Kit is for research use only and not for in vitro diagnostic use.
Customer Experience with the EasyScreen™ SARS-CoV-2 Detection Kit (RP012)
KH Labor GmbH, a provider of laboratory diagnostic services across multiple sites in Germany, and has been user of the Genetic Signatures’ SARS-CoV-2 Detection Kit (RP012) since the end of 2020. Genetic Signatures has developed a close partnership with Dr. Mark Wasner, Technical Head of Microbiology and Molecular Diagnostics, and his laboratory team. To support KH Labor’s high throughput requirements, the EasyScreen™ SARS-CoV-2 Detection Kit is run on Genetic Signatures GS-1000 automated sample processing system. The advantages of Genetic Signatures’ 3base™ technology gave KH Labor confidence in their ability to detect known and new SARS-CoV-2 variants. In addition, their workflow was further improved by the implementation of Genetic Signatures’ SARS-CoV-2 PCR FAST protocol, which significantly reduced the ‘time to result’ and improved daily throughput due to faster thermocycling.

References:
- Kevadiya, B.D., Machhi, J., Herskovitz, J. et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 20, 593–605 (2021). https://doi.org/10.1038/s41563-020-00906-zhttps://www.nature.com/articles/s41563-020-00906-z
- Liu, R. et al. Positive rate of RT–PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin. Chim. Acta 505, 172–175 (2020).
- Genetic Signatures. EasyScreen™ Respiratory Pathogen Detection, 2021, [online], accessed on 17/12/2021, https://geneticsignatures.com/au/our-products/respiratory/#Respiratory-Pathogens.
- Alsuliman T, Sulaiman R, Ismail S, Srour M, Alrstom A. COVID-19 paraclinical diagnostic tools: updates and future trends. Curr Res Transl Med. (2020) 68:83–91. doi: 10.1016/j.retram.2020.06.001
- Wu SY, Yau HS, Yu MY, Tsang HF, Chan LWC, Cho WCS, et al. The diagnostic methods in the COVID-19 pandemic, today and in the future. Expert Rev Mol Diagn. (2020) 20:985–93. doi: 10.1080/14737159.2020.1816171
- Shen, Z. et al. Genomic diversity of severe acute respiratory syndrome-coronavirus 2 in patients with coronavirus disease 2019. Clin. Infect. Dis. 71, 713–720 (2020).
- Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev Mol Diagn. (2020) 20:453–4. doi: 10.1080/14737159.2020.1757437
- U.S. Food and Drug Administration, 2021, SARS-CoV-2 Viral Mutations: Impact on COVID-19 Tests, [online], accessed on 12/10/2021, https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests
- World Health Organisation (WHO), 2021, Tracking SARS-CoV-2 variants, [online], accessed on 12/12/2021, https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
- GISAD, 2021, Tracking of Variants, [online], accessed on 12/12/2021, https://www.gisaid.org/hcov19-variants/ database
- World Health Organisation (WHO), 26 Nov 2021, Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. [online], accessed 12/12/2021, https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern
- ECDC, 11 Dec 2021, Epidemiological update: Omicron variant of concern (VOC) – data as of 11 December 2021 (12:00), [online], accessed 12/12/2021, https://www.ecdc.europa.eu/en/news-events/epidemiological-update-omicron-variant-concern-voc-data-11-december-2021
- Centers for Disease Control and Prevention (CDC), 13 Dec 2021, Omicron Variant: What you need to know, [online], accessed 12/12/2021, https://www.cdc.gov/coronavirus/2019-ncov/variants/omicron-variant.html
- UK Health Security Agency, 26 Nov 2021, SARS-CoV-2 variants of concern and variants under investigation in England, technical briefing 29 (PDF) (Briefing). Public Health England. 26 November 2021. GOV-10481. Archived (PDF) from the original on 27 November 2021, [online], accessed on 13/12/2021, https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1036501/Technical_Briefing_29_published_26_November_2021.pdf
- Katelyn Jetelina, 5 Dec 2021, Omicron Update: Dec 4. Your Local Epidemiologist, [online], accessed on 10/12/2021, https://yourlocalepidemiologist.substack.com/p/omicron-update-dec-4
- Juliet R.C. Pulliam, Cari van Schalkwyk, Nevashan Govender, Anne von Gottberg, Cheryl Cohen, Michelle J. Groome, Jonathan Dushoff, Koleka Mlisana, Harry Moultrie. medRxiv 2021.11.11.21266068; doi: https://doi.org/10.1101/2021.11.11.21266068
- ABC News, citing the Kirby Institute, 12/12/2021, [online], accessed on 12/12/2021, https://www.abc.net.au/news/2021-12-12/omicron-coronavirus-inside-the-research-lab-vaccines/100690964
- Ridhwaan Sulliman (Twitter), cited by Katelyn Jetelina, Dec 8, 2021, Omicron: We’re getting (some) answers, [online], accessed on 12/12/2021, https://yourlocalepidemiologist.substack.com/p/omicron-were-getting-some-answers
- Centers for Disease Control and Prevention (CDC), Dec 2021, What You Need to Know About Variants, [online], accessed on 12/12/2021, https://www.cdc.gov/coronavirus/2019-ncov/variants/about-variants.html
- Katelyn Jetelina, 13 Dec 2021, Omicron Update: Dec 13, [online], accessed on 15/12/2021, https://yourlocalepidemiologist.substack.com/p/omicron-update-dec-13
- Science Media Centre, 8 Dec 2021, Expert reaction to UKHSA update on Omicron risk assessment on S-gene target failure. Quotation of Dr Julian Tang, Honorary Associate Professor/Clinical Virologist, Respiratory Sciences, University of Leicester, [online], accessed on 12/12/2021, https://www.sciencemediacentre.org/expert-reaction-to-ukhsa-update-on-omicron-risk-assessment-and-s-gene-target-failure/
- Genetic Signatures, 11 Oct 2021, Validation of the EasyScreen 3base SARS-CoV-2 Detection Kit RP012 on Saliva Samples. NRL Workshop – 11-13th of October 2021, [online], accessed on 14/12/2021, https://geneticsignatures.com/au/resource/nrl-presentation-2021/
